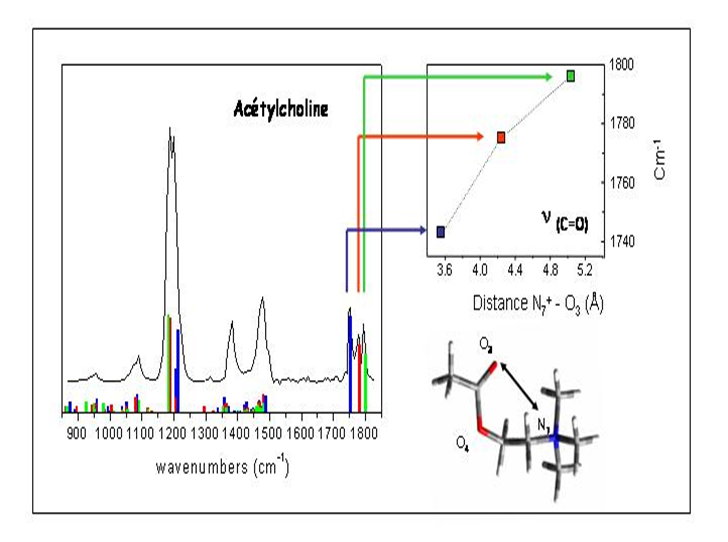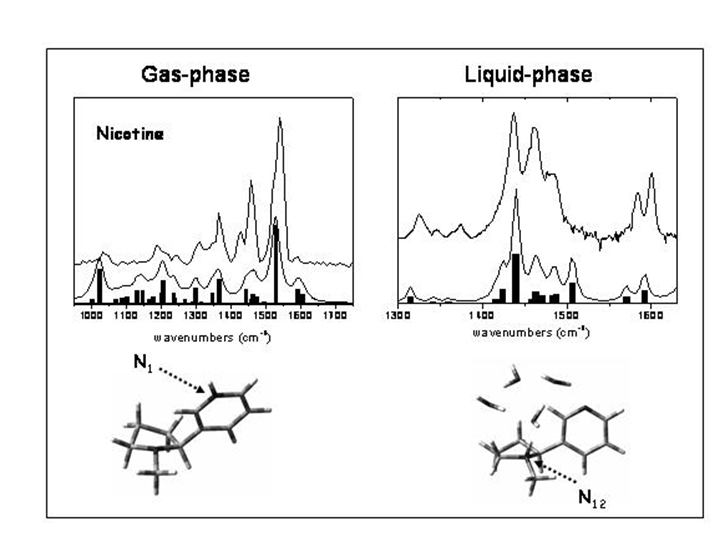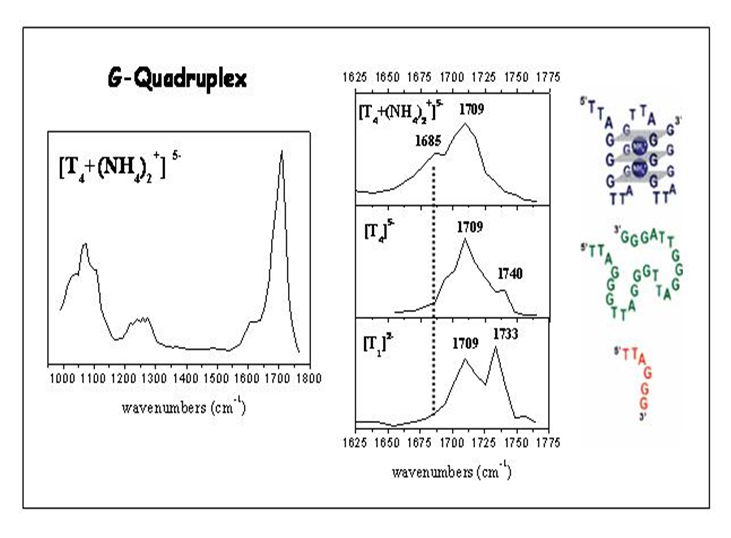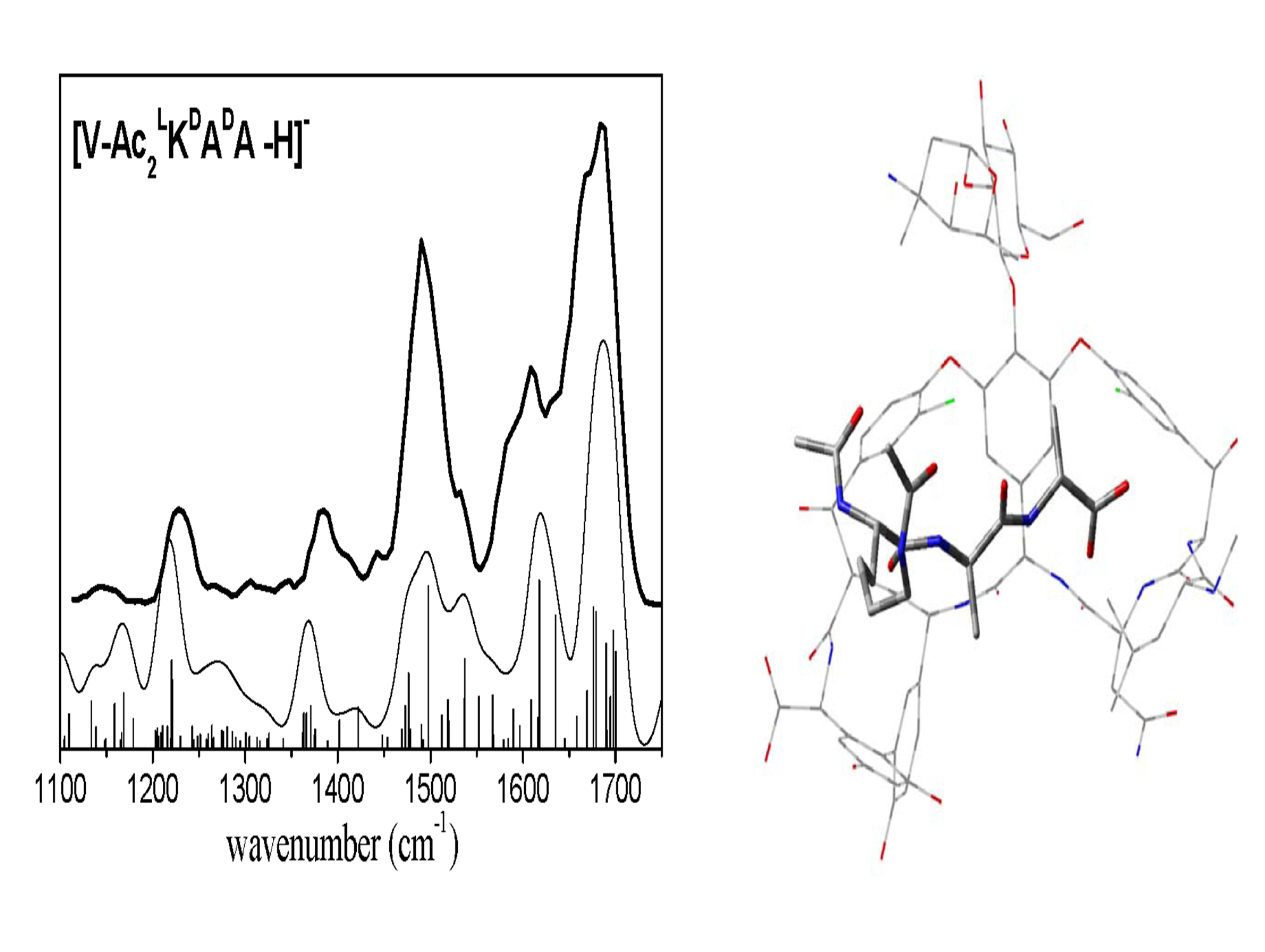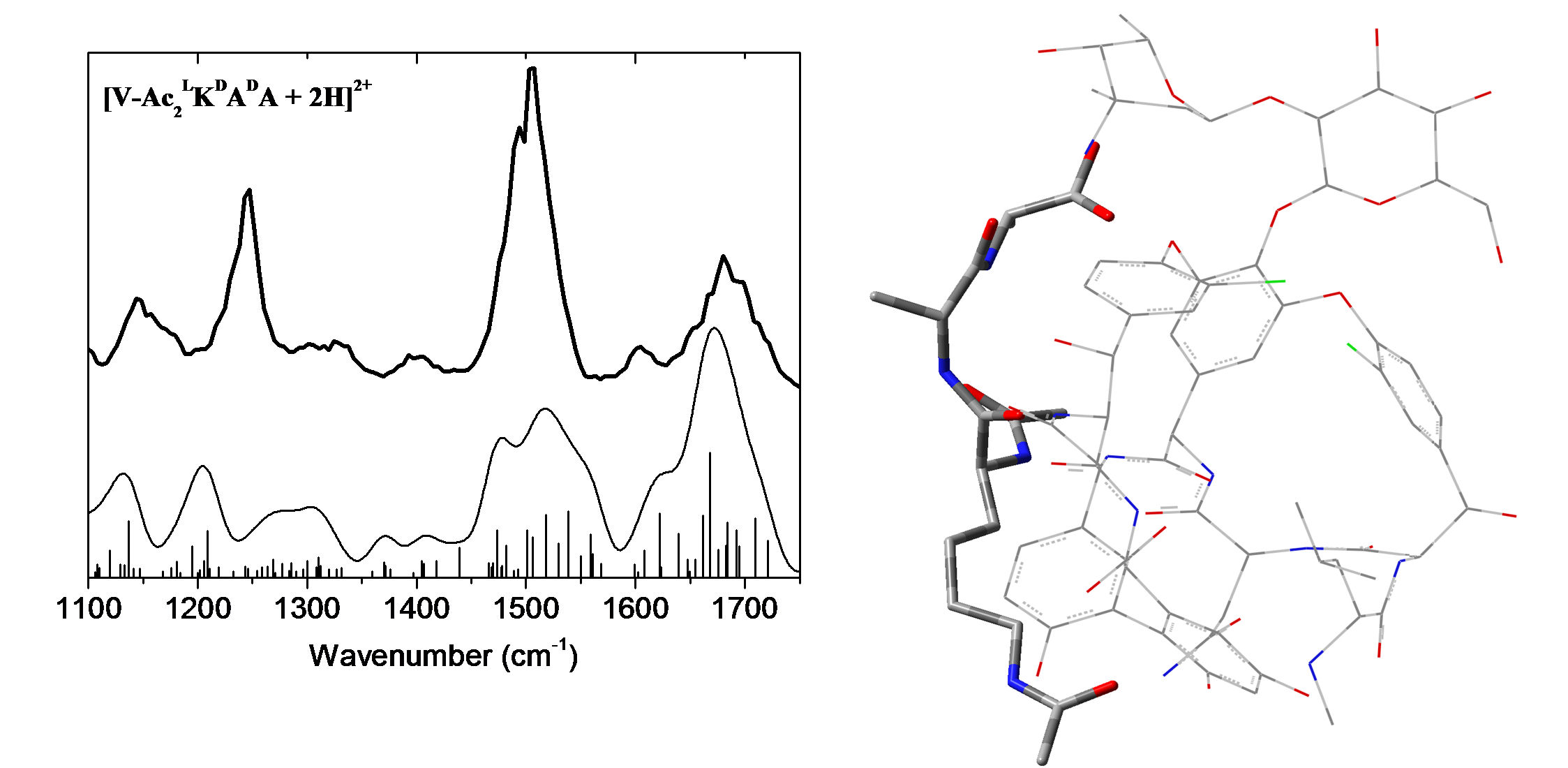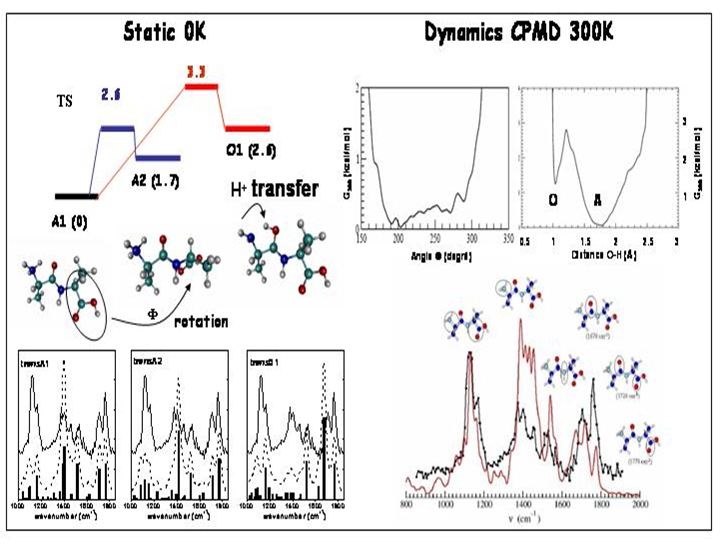
IRMPD offers a direct possibility for determination of protonation sites by comparison between experimental IR absorption spectrum and the predicted spectra of the different tautomers. In the case of protonated dialanine, static calculations of the lowest energy configurations show that the proton can be located at the N-terninal (labelled A1 and A2) or C=O (labelled O1) amide groups at 300 K. These conformations are separated by energy barriers with transition states accessible at room temperature. Conformations A1 and A2 give the best agreement with the experimental spectrum while conformation O1 contributes to a much less extent to the experimental spectrum.
Car-Parrinello molecular dynamics (CPMD) simulations on Ala-AlaH+ peptide at 300K have shown a continual and recurrent isomerization dynamics between the two A1 and A2 conformers, i.e. the internal energy of the peptide at 300K is sufficient to overcome the energy barrier separating the A1 and A2 basins on the potential energy surface. Besides, one important result is the observation of spontaneous and reversible proton transfer events between NH3+ (A conformer) and the adjacent C=O amide (O conformer). This indicates that O1 is a metastable state which is thermally accessible at 300K. The IR spectrum extracted from the CPMD simulation is calculated from the Fourier Transform of the dipole time correlation function. All pertinent band shaping effects, such as anharmonicity and temperature are incorporated ab inito via the CPMD methodology.